In the Pollack Laboratory, we study the physics of biomolecules: DNA, RNA and proteins. Although we rely a lot on small angle x-ray scattering (SAXS) experiments to understand these complex molecules, we also utilize complementary spectroscopic tools to full understand the workings of these molecules. Here we highlight several spectroscopic techniques to understand the biophysics of these molecules.
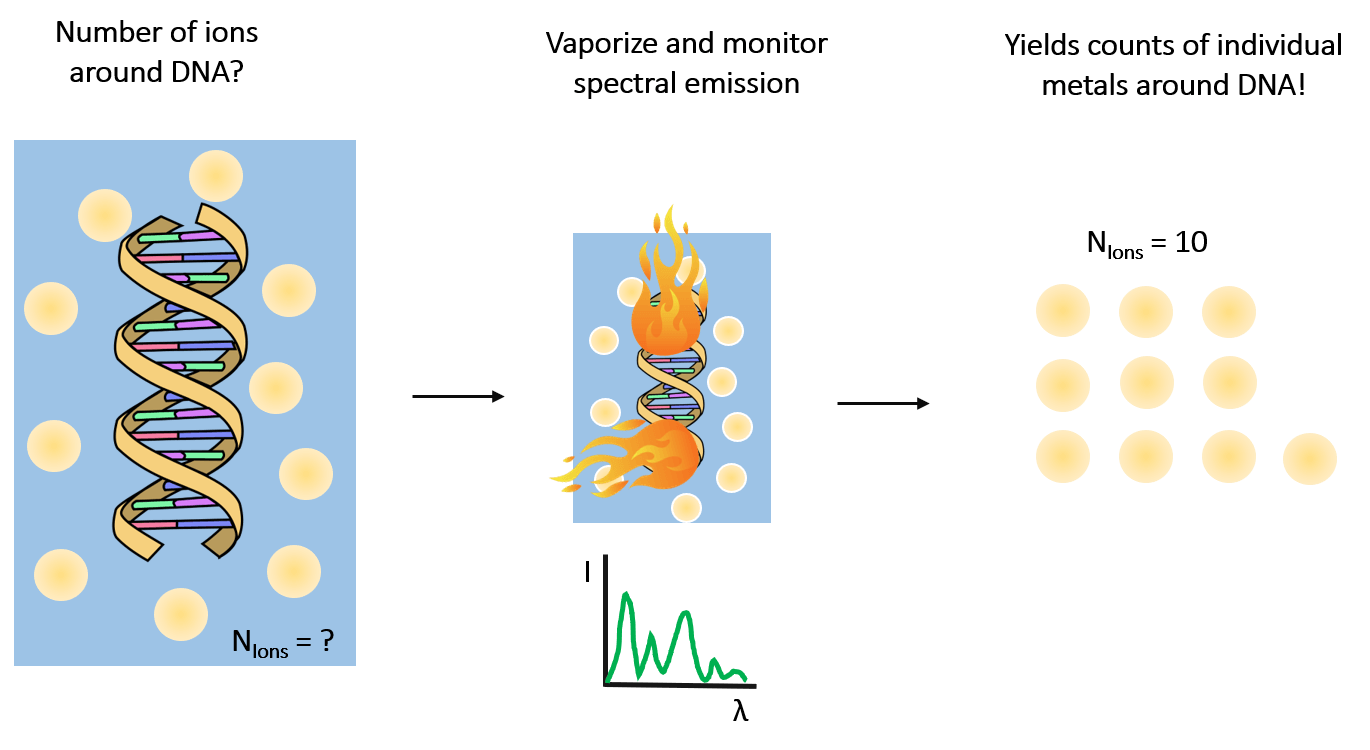
One ion, two ion, three ion, four… counting metals around nucleic acids
Biomolecules swim in a cellular soup of water, ions, cofactors and other machinery. Despite their simplicity, water and metal ions are tasked with folding more complicated biomolecules into functioning structures. Nucleic acids (such as DNA and RNA) are charged polymers and interact with cell ions in numerous exotic ways. Knowledge of exactly which, and how many metal ions are around a nucleic acid provides insight into how these molecules reach functioning structures and propels theoretical modeling.
In the lab, we routinely perform highly precise ion counting measurements to resolve this information using a technique known as atomic emission spectroscopy (illustrated in the panel below). By vaporizing our sample of interest, we detect spectral lines at distinct wavelengths that allow quantification of the identity and number of ions in the sample [1].
We recently counted the number of magnesium ions around a given RNA and found (6.34±0.08) of them [2]!
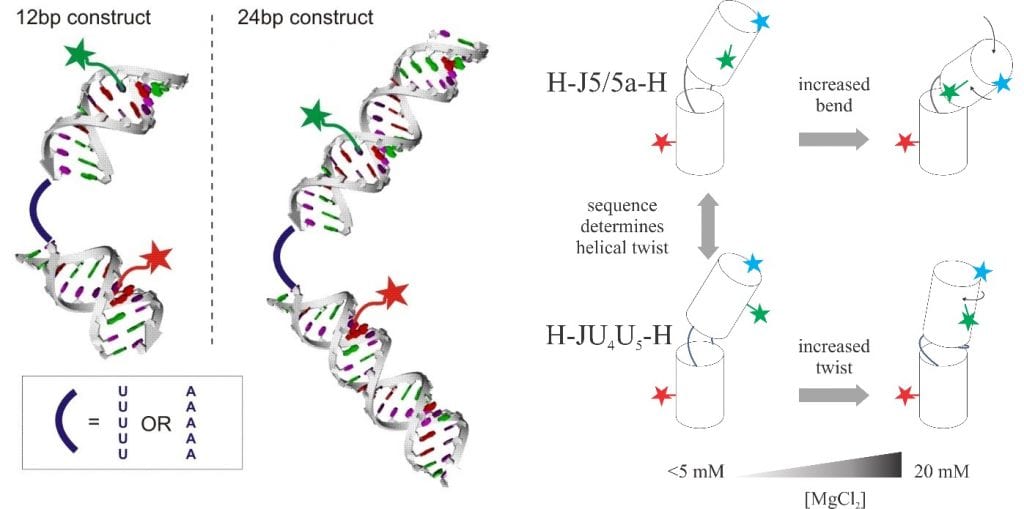
Using Single-Molecule Fluorescence Spectroscopy to look at RNA folding
RNAs are flexible molecules and their flexibility can be tuned by the internal structures, sequences, and solvent conditions [3]. Knowledge of how conformations of simple RNA constructs respond to different ionic strengths can be generalized to complex functional RNA molecules and their folding energy landscape [4]. The technique of single-molecule fluorescent energy transfer serves as a molecular ruler to measure the distance between the fluorophores attached to two spots of an RNA molecule. By exploiting different labeling positions, we can break the degeneracy of twist and bend as a function of ion strengths.
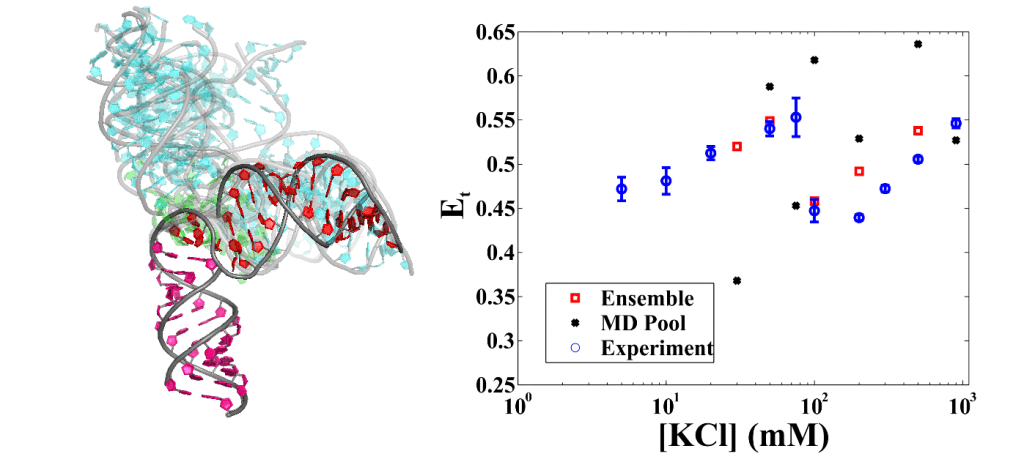
Using combination of smFRET, SAXS and EOM to study RNA conformational ensembles
SAXS and EOM (ensemble optimization method) combined form a powerful tool to reveal RNA conformational ensembles in vitro with the help of unbiased molecular dynamics simulations. Ensembles of flexible RNA constructs under varying solution environment were found [5]. With knowledge that smFRET is a binomial process and that different conformational states have different contributions to the empirical energy transfer rate, we validated the SAXS-derived ensemble by carrying out sampling simulations. To our surprise, we found that a portion of the flexible RNA molecules are elongated at high ionic strength.
References
- Plumridge et al. The impact of base stacking on the conformations and electrostatics of single-stranded DNA. Nucleic Acids Research, 2017. doi: 10.1093/nar/gkx140
- Plumridge et al. Sequence drives unique conformational and electrostatic signatures of single-stranded RNA. In press.
- Sutton at al. Tuning RNA Flexibility with Helix Length and Junction Sequence. Biophysical Journal, 2015. doi: 10.1016/j.bpj.2015.10.039
- Chen et al. How the Conformations of an Internal Junction Contribute to Fold an RNA Domain, The Journal of Physical Chemistry B 2018. doi: 10.1021/acs.jpcb.8b07262
- Chen et al. Conformations of an RNA Helix-Junction-Helix Construct Revealed by SAXS Refinement of MD Simulations. Biophysical Journal 2019. doi: 10.1016/J.BPJ.2018.11.020